Processing data with pandas II#
Note
It is not recommended to use Binder for this lesson.
This week we will continue developing our skills using pandas to process real data.
Motivation#
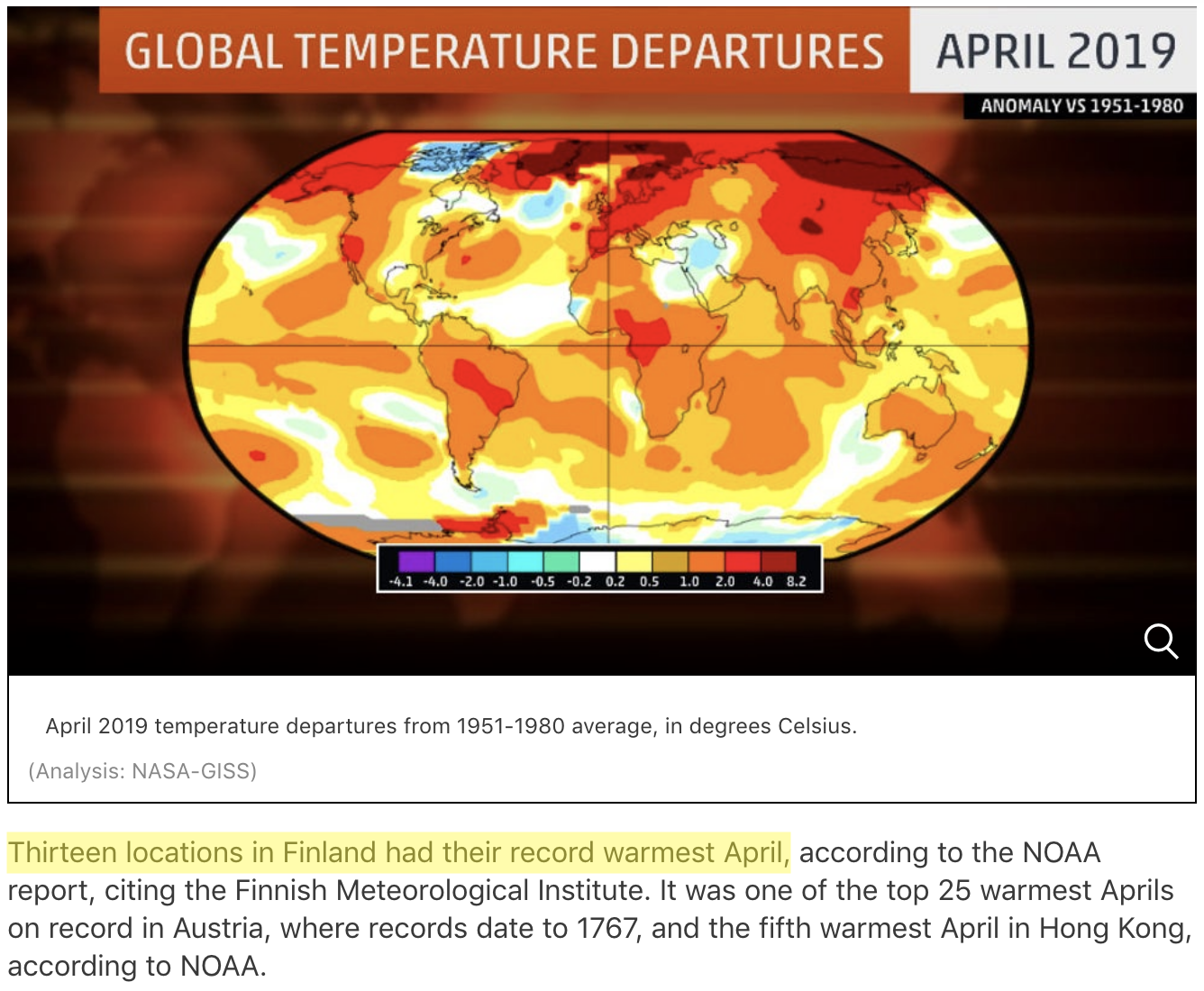 Source: https://weather.com/news/climate/news/2019-05-20-april-2019-global-temperatures-nasa-noaa
Source: https://weather.com/news/climate/news/2019-05-20-april-2019-global-temperatures-nasa-noaa
April 2019 was the second warmest April on record globally, and the warmest on record at 13 weather stations in Finland. In this lesson, we will use our data manipulation and analysis skills to analyze weather data, and investigate the claim that April 2019 was the warmest on record across Finland.
Along the way we will cover a number of useful techniques in pandas including:
renaming columns
iterating data frame rows and applying functions
data aggregation
repeating the analysis task for several input files
Input data#
In the lesson this week we are using weather observation data from Finland downloaded from NOAA. You will be working with data from a total of 15 different weather observation stations in Finland.
Downloading the data#
Note
The instructions about downloading the data only apply to students working with Jupyter Lab installed on their own computers. If you are using the CSC Notebooks you do not need follow these steps!
If you are working on a Jupyter environment installed on your own computer (i.e., not the CSC notebooks), the first step for today’s lesson is to get the data. We recommend using the command line tool wget for downloading the data. wget is hopefully already installed on your computer.
First, you need to open a new terminal window in Jupyter Lab (from File -> New -> Terminal). Once the terminal window is open, you will need to navigate to the L6 directory:
cd path/to/L6/
where path/to/ should be replaced with the correct path for the Lesson 6 materials on your computer. Once in the correct directory, you can confirm this by typing:
ls
You should see something like the following output:
advanced-data-processing-with-pandas.ipynb errors.ipynb img
debugging.ipynb gcp-5-assertions.ipynb
Next, you can download the data files using wget:
wget https://davewhipp.github.io/data/Finland-weather-data-full.tar.gz
After the download completes, you can extract the data files using the tar command:
tar zxvf Finland-weather-data-full.tar.gz
At this stage you should have a new directory called data that contains the data for this week’s lesson. You can confirm this by listing the contents of the data-folder:
ls data
You should see something like the following:
028360.txt 029070.txt 029440.txt 029740.txt 6367598020644inv.txt
028690.txt 029110.txt 029500.txt 029810.txt 6367598020644stn.txt
028750.txt 029170.txt 029700.txt 029820.txt
028970.txt 029350.txt 029720.txt 3505doc.txt
Now you should be all set to proceed with the lesson!
About the data#
As part of the download there are a number of files that describe the weather data. These metadata files include:
A list of stations: 6367598020644stn.txt
Details about weather observations at each station: 6367598020644inv.txt
A data description (i.e., column names): 3505doc.txt
The input data for this week are separated with varying number of spaces (i.e., fixed width). The first lines and columns of the data look like following:
USAF WBAN YR--MODAHRMN DIR SPD GUS CLG SKC L M H VSB MW MW MW MW AW AW AW AW W TEMP DEWP SLP ALT STP MAX MIN PCP01 PCP06 PCP24 PCPXX SD
029440 99999 190601010600 090 7 *** *** OVC * * * 0.0 ** ** ** ** ** ** ** ** * 27 **** 1011.0 ***** ****** *** *** ***** ***** ***** ***** **
029440 99999 190601011300 *** 0 *** *** OVC * * * 0.0 ** ** ** ** ** ** ** ** * 27 **** 1015.5 ***** ****** *** *** ***** ***** ***** ***** **
029440 99999 190601012000 *** 0 *** *** OVC * * * 0.0 ** ** ** ** ** ** ** ** * 25 **** 1016.2 ***** ****** *** *** ***** ***** ***** ***** **
029440 99999 190601020600 *** 0 *** *** CLR * * * 0.0 ** ** ** ** ** ** ** ** * 26 **** 1016.2 ***** ****** *** *** ***** ***** ***** ***** **
We will develop our analysis workflow using data for a single station. Then, we will repeat the same process for all the stations.
Reading the data#
In order to get started, let’s first import pandas:
import pandas as pd
At this point, we can already have a quick look at the data file 029440.txt for Tampere Pirkkala and how it is structured. We can notice at least two things we need to consider when reading in the data:
Input data structure
Delimiter: The data are separated with a varying amount of spaces. If you check out the documentation for the read_csv() method, you can see that there are two different ways of doing this. We can use either
sep='\s+'ordelim_whitespace=True(but not both at the same time). In this case, we prefer to usedelim_whitespaceparameter.No Data values: No data values in the NOAA data are coded with varying number of
*. We can tell pandas to consider those characters as NaNs by specifyingna_values=['*', '**', '***', '****', '*****', '******'].
# Define absolute path to the file
fp = r"/home/jovyan/shared/data/L6/029440.txt"
Show code cell content
# IGNORE THIS CELL
# It is only needed to generate the course website
fp = r"data/029440.txt"
# Read data using varying amount of spaces as separator and specifying * characters as NoData values
data = pd.read_csv(
fp, delim_whitespace=True, na_values=["*", "**", "***", "****", "*****", "******"]
)
/var/folders/lp/cjwc88bd3w10sg327y_4ghg0fsk7jj/T/ipykernel_42174/972691635.py:2: DtypeWarning: Columns (29,30,31) have mixed types. Specify dtype option on import or set low_memory=False.
data = pd.read_csv(
Note
If you are working with Jupyter Lab installed locally on your own computer and extracted your files in the L6 notebooks directory, then you can use the file path below.
# Define relative path to the file
fp = r"data/029440.txt"
Let’s see how the data looks by printing the first five rows with the head() function:
data.head()
| USAF | WBAN | YR--MODAHRMN | DIR | SPD | GUS | CLG | SKC | L | M | ... | SLP | ALT | STP | MAX | MIN | PCP01 | PCP06 | PCP24 | PCPXX | SD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 29440 | 99999 | 190601010600 | 90.0 | 7.0 | NaN | NaN | OVC | NaN | NaN | ... | 1011.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 1 | 29440 | 99999 | 190601011300 | NaN | 0.0 | NaN | NaN | OVC | NaN | NaN | ... | 1015.5 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2 | 29440 | 99999 | 190601012000 | NaN | 0.0 | NaN | NaN | OVC | NaN | NaN | ... | 1016.2 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | 29440 | 99999 | 190601020600 | NaN | 0.0 | NaN | NaN | CLR | NaN | NaN | ... | 1016.2 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | 29440 | 99999 | 190601021300 | 270.0 | 7.0 | NaN | NaN | OVC | NaN | NaN | ... | 1015.6 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
5 rows × 33 columns
All seems ok. However, we won’t be needing all of the 33 columns for detecting warm temperatures in April. We can check all column names by running data.columns:
data.columns
Index(['USAF', 'WBAN', 'YR--MODAHRMN', 'DIR', 'SPD', 'GUS', 'CLG', 'SKC', 'L',
'M', 'H', 'VSB', 'MW', 'MW.1', 'MW.2', 'MW.3', 'AW', 'AW.1', 'AW.2',
'AW.3', 'W', 'TEMP', 'DEWP', 'SLP', 'ALT', 'STP', 'MAX', 'MIN', 'PCP01',
'PCP06', 'PCP24', 'PCPXX', 'SD'],
dtype='object')
A description for all these columns is available in the metadata file data/3505doc.txt.
Reading in the data once again#
This time, we will read in only some of the columns using the usecols parameter. Let’s read in columns that might be somehow useful to our analysis, or at least that contain some values that are meaningful to us, including the station name, timestamp, and data about wind and temperature: 'USAF','YR--MODAHRMN', 'DIR', 'SPD', 'GUS','TEMP', 'MAX', 'MIN'
# Read in only selected columns
data = pd.read_csv(
fp,
delim_whitespace=True,
usecols=["USAF", "YR--MODAHRMN", "DIR", "SPD", "GUS", "TEMP", "MAX", "MIN"],
na_values=["*", "**", "***", "****", "*****", "******"],
)
# Check the dataframe
data.head()
| USAF | YR--MODAHRMN | DIR | SPD | GUS | TEMP | MAX | MIN | |
|---|---|---|---|---|---|---|---|---|
| 0 | 29440 | 190601010600 | 90.0 | 7.0 | NaN | 27.0 | NaN | NaN |
| 1 | 29440 | 190601011300 | NaN | 0.0 | NaN | 27.0 | NaN | NaN |
| 2 | 29440 | 190601012000 | NaN | 0.0 | NaN | 25.0 | NaN | NaN |
| 3 | 29440 | 190601020600 | NaN | 0.0 | NaN | 26.0 | NaN | NaN |
| 4 | 29440 | 190601021300 | 270.0 | 7.0 | NaN | 27.0 | NaN | NaN |
Okay so we can see that the data was successfully read to the DataFrame and we also seemed to be able to convert the asterisk (*) characters into NaN values.
Renaming columns#
As we saw above some of the column names are a bit awkward and difficult to interpret. Luckily, it is easy to alter labels in a pandas DataFrame using the rename function. In order to change the column names, we need to tell pandas how we want to rename the columns using a dictionary that lists old and new column names
Let’s first check again the current column names in our DataFrame:
data.columns
Index(['USAF', 'YR--MODAHRMN', 'DIR', 'SPD', 'GUS', 'TEMP', 'MAX', 'MIN'], dtype='object')
Dictionaries
A dictionary is a specific type of data structure in Python for storing key-value pairs. In this course, we will use dictionaries mainly when renaming columns in a pandas DataFrame, but dictionaries are useful for many different purposes! For more information about Python dictionaries, check out this tutorial.
We can define the new column names using a dictionary where we list “key: value” pairs, in which the original column name (the one which will be replaced) is the key and the new column name is the value.
Let’s change the following:
YR--MODAHRMNtoTIMESPDtoSPEEDGUStoGUST
# Create the dictionary with old and new names
new_names = {"YR--MODAHRMN": "TIME", "SPD": "SPEED", "GUS": "GUST"}
# Let's see what the variable new_names look like
new_names
{'YR--MODAHRMN': 'TIME', 'SPD': 'SPEED', 'GUS': 'GUST'}
# Check the data type of the new_names variable
type(new_names)
dict
From above we can see that we have successfully created a new dictionary.
Now we can change the column names by passing that dictionary using the parameter columns in the rename() function:
# Rename the columns
data = data.rename(columns=new_names)
# Print the new columns
print(data.columns)
Index(['USAF', 'TIME', 'DIR', 'SPEED', 'GUST', 'TEMP', 'MAX', 'MIN'], dtype='object')
Perfect, now our column names are easier to understand and use.
Check your understanding#
The temperature values in our data files are again in Fahrenheit. As you might guess, we will soon convert these temperatures in to Celsius. In order to avoid confusion with the columns, let’s rename the column TEMP to TEMP_F. Let’s also rename USAF to STATION_NUMBER.
Show code cell content
# Solution
# Create the dictionary with old and new names
new_names = {"USAF": "STATION_NUMBER", "TEMP": "TEMP_F"}
# Rename the columns
data = data.rename(columns=new_names)
# Check the output
data.head()
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | |
|---|---|---|---|---|---|---|---|---|
| 0 | 29440 | 190601010600 | 90.0 | 7.0 | NaN | 27.0 | NaN | NaN |
| 1 | 29440 | 190601011300 | NaN | 0.0 | NaN | 27.0 | NaN | NaN |
| 2 | 29440 | 190601012000 | NaN | 0.0 | NaN | 25.0 | NaN | NaN |
| 3 | 29440 | 190601020600 | NaN | 0.0 | NaN | 26.0 | NaN | NaN |
| 4 | 29440 | 190601021300 | 270.0 | 7.0 | NaN | 27.0 | NaN | NaN |
Data properties#
As we learned last week, it’s always a good idea to check basic properties of the input data before proceeding with the data analysis. Let’s check the:
Number of rows and columns
data.shape
(757983, 8)
Top and bottom rows
data.head()
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | |
|---|---|---|---|---|---|---|---|---|
| 0 | 29440 | 190601010600 | 90.0 | 7.0 | NaN | 27.0 | NaN | NaN |
| 1 | 29440 | 190601011300 | NaN | 0.0 | NaN | 27.0 | NaN | NaN |
| 2 | 29440 | 190601012000 | NaN | 0.0 | NaN | 25.0 | NaN | NaN |
| 3 | 29440 | 190601020600 | NaN | 0.0 | NaN | 26.0 | NaN | NaN |
| 4 | 29440 | 190601021300 | 270.0 | 7.0 | NaN | 27.0 | NaN | NaN |
data.tail()
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | |
|---|---|---|---|---|---|---|---|---|
| 757978 | 29440 | 201910012220 | 130.0 | 3.0 | NaN | 39.0 | NaN | NaN |
| 757979 | 29440 | 201910012250 | 110.0 | 3.0 | NaN | 37.0 | NaN | NaN |
| 757980 | 29440 | 201910012300 | 100.0 | 2.0 | NaN | 38.0 | NaN | NaN |
| 757981 | 29440 | 201910012320 | 100.0 | 3.0 | NaN | 37.0 | NaN | NaN |
| 757982 | 29440 | 201910012350 | 110.0 | 3.0 | NaN | 37.0 | NaN | NaN |
Data types of the columns
data.dtypes
STATION_NUMBER int64
TIME int64
DIR float64
SPEED float64
GUST float64
TEMP_F float64
MAX float64
MIN float64
dtype: object
Descriptive statistics
data.describe()
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | |
|---|---|---|---|---|---|---|---|---|
| count | 757983.0 | 7.579830e+05 | 699256.000000 | 750143.000000 | 19906.000000 | 754862.000000 | 23869.000000 | 23268.000000 |
| mean | 29440.0 | 1.999974e+11 | 233.499846 | 6.742641 | 20.147996 | 40.409778 | 45.373539 | 35.783737 |
| std | 0.0 | 1.629544e+09 | 209.707258 | 4.296191 | 7.415138 | 17.898715 | 18.242679 | 17.195427 |
| min | 29440.0 | 1.906010e+11 | 10.000000 | 0.000000 | 11.000000 | -33.000000 | -26.000000 | -32.000000 |
| 25% | 29440.0 | 1.989083e+11 | 130.000000 | 3.000000 | 14.000000 | 29.000000 | 32.000000 | 26.000000 |
| 50% | 29440.0 | 2.004042e+11 | 200.000000 | 7.000000 | 18.000000 | 39.000000 | 44.000000 | 36.000000 |
| 75% | 29440.0 | 2.012050e+11 | 270.000000 | 9.000000 | 26.000000 | 54.000000 | 60.000000 | 49.000000 |
| max | 29440.0 | 2.019100e+11 | 990.000000 | 61.000000 | 108.000000 | 91.000000 | 91.000000 | 81.000000 |
Here we can see that there are varying number of observations per column (look at the count row above), because some of the columns have missing values.
Using your own functions in pandas#
Now it’s again time to convert temperatures from Fahrenheit to Celsius! Yes, we have already done this many times before, but this time we will learn how to apply our own functions to data in a pandas DataFrame.
First, we will define a function for the temperature conversion, and then we will apply this function for each Fahrenheit value on each row of the DataFrame. The output celsius values will be stored in a new column called TEMP_C.
To begin we will see how we can apply the function row-by-row using a for loop and then we will learn how to apply the function to all rows more efficiently all at once.
Defining the function#
For both of these approaches, we first need to define our function to convert temperature from Fahrenheit to Celsius.
def fahr_to_celsius(temp_fahrenheit):
"""Function to convert Fahrenheit temperature into Celsius.
Parameters
----------
temp_fahrenheit: int | float
Input temperature in Fahrenheit (should be a number)
Returns
-------
Temperature in Celsius (float)
"""
# Convert the Fahrenheit into Celsius
converted_temp = (temp_fahrenheit - 32) / 1.8
return converted_temp
To make sure everything is working properly, let’s test the function with a known value.
fahr_to_celsius(32)
0.0
Let’s also print out the first rows of our data frame to see our input data before further processing.
data.head()
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | |
|---|---|---|---|---|---|---|---|---|
| 0 | 29440 | 190601010600 | 90.0 | 7.0 | NaN | 27.0 | NaN | NaN |
| 1 | 29440 | 190601011300 | NaN | 0.0 | NaN | 27.0 | NaN | NaN |
| 2 | 29440 | 190601012000 | NaN | 0.0 | NaN | 25.0 | NaN | NaN |
| 3 | 29440 | 190601020600 | NaN | 0.0 | NaN | 26.0 | NaN | NaN |
| 4 | 29440 | 190601021300 | 270.0 | 7.0 | NaN | 27.0 | NaN | NaN |
Iterating over rows#
We can apply the function one row at a time using a for loop and the iterrows() method. This will allow us to iterate row by row using iterrows() in a for loop to repeat a given process for each row in a pandas DataFrame. Please note that iterating over rows is a rather inefficient approach, but it is still useful to understand the logic behind the iteration.
When using the iterrows() method it is important to understand that iterrows() accesses not only the values of one row, but also the index of the row as well.
Let’s start with a simple for loop that goes through each row in our DataFrame.
Note
We use single quotes to select the column TEMP_F of the row in the example below. This is because using double quotes would result in a SyntaxError since Python would interpret this as the end of the string for the print() function.
# Iterate over the rows
for idx, row in data.iterrows():
# Print the index value
print(f"Index: {idx}")
# Print the row
print(f"Temp F: {row['TEMP_F']}\n")
break
Index: 0
Temp F: 27.0
Breaking a loop
When developing code in a for loop, you do not always need to go through the entire loop in order to test things out.
The break statement in Python terminates the current loop whereever it is placed and we can use it here just to check out the values on the first row (based on the first iteration in the for loop.
This can be helpful when working with a large data file or dataset, because you might not want to print thousands of values to the screen!
We can see that the idx variable indeed contains the index value at position 0 (the first row) and the row variable contains all the data from that given row stored as a pandas Series.
Let’s now create an empty column TEMP_C for the Celsius temperatures and update the values in that column using the fahr_to_celsius function we defined earlier.
# Create an empty float column for the output values
data["TEMP_C"] = 0.0
# Iterate over the rows
for idx, row in data.iterrows():
# Convert the Fahrenheit to Celsius
celsius = fahr_to_celsius(row["TEMP_F"])
# Update the value of 'Celsius' column with the converted value
data.at[idx, "TEMP_C"] = celsius
Reminder: .at or .loc?
Here, you could also use data.loc[idx, new_column] = celsius to achieve the same result.
If you only need to access a single value in a DataFrame, DataFrame.at is faster than DataFrame.loc, which is designed for accessing groups of rows and columns.
Finally, let’s see how our DataFrame looks like now after the calculations above.
data.head()
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | TEMP_C | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 29440 | 190601010600 | 90.0 | 7.0 | NaN | 27.0 | NaN | NaN | -2.777778 |
| 1 | 29440 | 190601011300 | NaN | 0.0 | NaN | 27.0 | NaN | NaN | -2.777778 |
| 2 | 29440 | 190601012000 | NaN | 0.0 | NaN | 25.0 | NaN | NaN | -3.888889 |
| 3 | 29440 | 190601020600 | NaN | 0.0 | NaN | 26.0 | NaN | NaN | -3.333333 |
| 4 | 29440 | 190601021300 | 270.0 | 7.0 | NaN | 27.0 | NaN | NaN | -2.777778 |
Applying the function#
pandas DataFrames and Series have a dedicated method .apply() for applying functions on columns (or rows!). When using .apply(), we pass the function name (without parentheses!) as an argument to the apply() method. Let’s start by applying the function to the TEMP_F column that contains the temperature values in Fahrenheit.
data["TEMP_F"].apply(fahr_to_celsius)
0 -2.777778
1 -2.777778
2 -3.888889
3 -3.333333
4 -2.777778
...
757978 3.888889
757979 2.777778
757980 3.333333
757981 2.777778
757982 2.777778
Name: TEMP_F, Length: 757983, dtype: float64
The results look logical, so we can store them permanently into the TEMP_C column (overwriting the old values).
data["TEMP_C"] = data["TEMP_F"].apply(fahr_to_celsius)
We can also apply the function on several columns at once. Furthermore, we can re-order the dataframe at the same time.
data[["TEMP_F", "MIN", "MAX"]].apply(fahr_to_celsius)
| TEMP_F | MIN | MAX | |
|---|---|---|---|
| 0 | -2.777778 | NaN | NaN |
| 1 | -2.777778 | NaN | NaN |
| 2 | -3.888889 | NaN | NaN |
| 3 | -3.333333 | NaN | NaN |
| 4 | -2.777778 | NaN | NaN |
| ... | ... | ... | ... |
| 757978 | 3.888889 | NaN | NaN |
| 757979 | 2.777778 | NaN | NaN |
| 757980 | 3.333333 | NaN | NaN |
| 757981 | 2.777778 | NaN | NaN |
| 757982 | 2.777778 | NaN | NaN |
757983 rows × 3 columns
Check your understanding#
Convert 'TEMP_F', 'MIN', 'MAX' to Celsius by applying the function like we did above and store the outputs to new columns 'TEMP_C', 'MIN_C', 'MAX_C'.
Show code cell content
# Solution
data[["TEMP_C", "MIN_C", "MAX_C"]] = data[["TEMP_F", "MIN", "MAX"]].apply(
fahr_to_celsius
)
Note
Applying the function on all columns using data.apply(fahr_to_celsius) would not give an error for the data we are using, but the results also do not make much sense for columns where input data are something other than Fahrenheit temperatures.
You might also notice that our conversion function would also allow us to
pass one column or the entire dataframe as a parameter. For example, like this: fahr_to_celsius(data["TEMP_F"]). However, the code is perhaps easier to follow when using the apply method.
Let’s check now take a look at the DataFrame contents.
data.head(10)
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | TEMP_C | MIN_C | MAX_C | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 29440 | 190601010600 | 90.0 | 7.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN |
| 1 | 29440 | 190601011300 | NaN | 0.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN |
| 2 | 29440 | 190601012000 | NaN | 0.0 | NaN | 25.0 | NaN | NaN | -3.888889 | NaN | NaN |
| 3 | 29440 | 190601020600 | NaN | 0.0 | NaN | 26.0 | NaN | NaN | -3.333333 | NaN | NaN |
| 4 | 29440 | 190601021300 | 270.0 | 7.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN |
| 5 | 29440 | 190601022000 | NaN | 0.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN |
| 6 | 29440 | 190601030600 | 270.0 | 7.0 | NaN | 26.0 | NaN | NaN | -3.333333 | NaN | NaN |
| 7 | 29440 | 190601031300 | 270.0 | 7.0 | NaN | 25.0 | NaN | NaN | -3.888889 | NaN | NaN |
| 8 | 29440 | 190601032000 | 270.0 | 7.0 | NaN | 24.0 | NaN | NaN | -4.444444 | NaN | NaN |
| 9 | 29440 | 190601040600 | NaN | 0.0 | NaN | 18.0 | NaN | NaN | -7.777778 | NaN | NaN |
Should I use .iterrows() or .apply()?
We are teaching the .iterrows() method because it helps to understand the structure of a DataFrame and the process of looping through DataFrame rows. However, using .apply() is often more efficient in terms of execution time.
At this point, the most important thing is that you understand what happens when you are modifying the values in a pandas DataFrame. When doing the course exercises, either of these approaches is ok!
Parsing dates#
As part of this lesson, we eventually want to group our data based on month in order to see whether the temperatures in April of 2019 were higher than average. Currently, the date and time information is stored in the column TIME (which was originally titled YR--MODAHRMN:
YR--MODAHRMN = YEAR-MONTH-DAY-HOUR-MINUTE IN GREENWICH MEAN TIME (GMT)
Let’s have a closer look at the date and time information we have by checking the values in that column, and their data type.
data["TIME"].head(10)
0 190601010600
1 190601011300
2 190601012000
3 190601020600
4 190601021300
5 190601022000
6 190601030600
7 190601031300
8 190601032000
9 190601040600
Name: TIME, dtype: int64
data["TIME"].tail(10)
757973 201910012050
757974 201910012100
757975 201910012120
757976 201910012150
757977 201910012200
757978 201910012220
757979 201910012250
757980 201910012300
757981 201910012320
757982 201910012350
Name: TIME, dtype: int64
The TIME column contains several observations per day (and even several observations per hour). The timestamp for the first observation is 190601010600, i.e. from 1st of January 1906 (way back!), and the timestamp for the latest observation is 201910012350.
data["TIME"].dtypes
dtype('int64')
The information in this column is stored as integer values.
We now want to aggregate the data on a monthly level, and in order to do so we need to “label” each row of data based on the month when the record was observed. In order to do this, we need to somehow separate information about the year and month for each row.
We can create these “labels” by making a new column containing information about the month (including the year, but excluding day, hours, and minutes).
Before further taking that step, we should first convert the contents in the TIME column to a new column with character string values for convenience.
# Convert to string
data["TIME_STR"] = data["TIME"].astype(str)
String slicing#
Now that we have converted the date and time information into character strings, we next need to “cut” the needed information from the string objects. If we look at the latest time stamp in the data (201910012350), you can see that there is a systematic pattern YEAR-MONTH-DAY-HOUR-MINUTE. Four first characters represent the year, and six first characters are year and month!
date = "201910012350"
date[0:6]
'201910'
Based on this information, we can slice the correct range of characters from the TIME_STR column using the pandas.Series.str.slice() method.
# Slice the string
data["YEAR_MONTH"] = data["TIME_STR"].str.slice(start=0, stop=6)
# Let's see what we have
data.head()
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | TEMP_C | MIN_C | MAX_C | TIME_STR | YEAR_MONTH | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 29440 | 190601010600 | 90.0 | 7.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN | 190601010600 | 190601 |
| 1 | 29440 | 190601011300 | NaN | 0.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN | 190601011300 | 190601 |
| 2 | 29440 | 190601012000 | NaN | 0.0 | NaN | 25.0 | NaN | NaN | -3.888889 | NaN | NaN | 190601012000 | 190601 |
| 3 | 29440 | 190601020600 | NaN | 0.0 | NaN | 26.0 | NaN | NaN | -3.333333 | NaN | NaN | 190601020600 | 190601 |
| 4 | 29440 | 190601021300 | 270.0 | 7.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN | 190601021300 | 190601 |
Nice! Now we have “labeled” the rows based on information about date and time, but only including the year and month in the labels.
Check your understanding#
Create a new column called 'MONTH' with information about the month of each observation, but excluding the year.
Show code cell content
# Solution
# Extract information about month from the TIME_STR column into a new column 'MONTH':
data["MONTH"] = data["TIME_STR"].str.slice(start=4, stop=6)
# Check the result
data[["YEAR_MONTH", "MONTH"]]
| YEAR_MONTH | MONTH | |
|---|---|---|
| 0 | 190601 | 01 |
| 1 | 190601 | 01 |
| 2 | 190601 | 01 |
| 3 | 190601 | 01 |
| 4 | 190601 | 01 |
| ... | ... | ... |
| 757978 | 201910 | 10 |
| 757979 | 201910 | 10 |
| 757980 | 201910 | 10 |
| 757981 | 201910 | 10 |
| 757982 | 201910 | 10 |
757983 rows × 2 columns
Datetime (optional for Lesson 6)#
In pandas, we can also convert dates and times into a new data type called datetime using the pandas.to_datetime function.
# Convert character strings to datetime
data["DATE"] = pd.to_datetime(data["TIME_STR"])
# Check the output
data["DATE"].head()
0 1906-01-01 06:00:00
1 1906-01-01 13:00:00
2 1906-01-01 20:00:00
3 1906-01-02 06:00:00
4 1906-01-02 13:00:00
Name: DATE, dtype: datetime64[ns]
Pandas Series datetime properties
There are several methods available for accessing information about the properties of datetime values. You can read more about datetime properties from the pandas documentation.
With the datetime column, we can now extract different time units using the pandas.Series.dt accessor.
data["DATE"].dt.year
0 1906
1 1906
2 1906
3 1906
4 1906
...
757978 2019
757979 2019
757980 2019
757981 2019
757982 2019
Name: DATE, Length: 757983, dtype: int64
data["DATE"].dt.month
0 1
1 1
2 1
3 1
4 1
..
757978 10
757979 10
757980 10
757981 10
757982 10
Name: DATE, Length: 757983, dtype: int64
We can also combine the datetime functionalities with other methods from pandas. For example, we can check the number of unique years in our input data:
data["DATE"].dt.year.nunique()
51
For the final analysis, we need combined information of the year and month. One way to achieve this is to use the format parameter to define the output datetime format according to strftime(format) method:
# Convert to datetime and keep only year and month
data["YEAR_MONTH_DT"] = pd.to_datetime(data["TIME_STR"], format="%Y%m", exact=False)
Note
exact=False finds the characters matching the specified format and drops out the rest (days, hours and minutes are excluded in the output).
data["YEAR_MONTH_DT"]
0 1906-01-01
1 1906-01-01
2 1906-01-01
3 1906-01-01
4 1906-01-01
...
757978 2019-10-01
757979 2019-10-01
757980 2019-10-01
757981 2019-10-01
757982 2019-10-01
Name: YEAR_MONTH_DT, Length: 757983, dtype: datetime64[ns]
Now we have a unique label for each month as a datetime object!
Aggregating data in pandas by grouping#
Here, we will learn how to use pandas.DataFrame.groupby, which is a handy method for compressing large amounts of data and computing statistics for subgroups.
In our case, we will use the groupby method to calculate the average temperatures for each month through these three steps:
Grouping the data based on the year and month
Calculating the average for each month (each group)
Storing those values into a new DataFrame called
monthly_data
Before we start grouping the data, let’s once again see what our data looks like.
print(f"number of rows: {len(data)}")
number of rows: 757983
data.head()
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | TEMP_C | MIN_C | MAX_C | TIME_STR | YEAR_MONTH | MONTH | DATE | YEAR_MONTH_DT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 29440 | 190601010600 | 90.0 | 7.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN | 190601010600 | 190601 | 01 | 1906-01-01 06:00:00 | 1906-01-01 |
| 1 | 29440 | 190601011300 | NaN | 0.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN | 190601011300 | 190601 | 01 | 1906-01-01 13:00:00 | 1906-01-01 |
| 2 | 29440 | 190601012000 | NaN | 0.0 | NaN | 25.0 | NaN | NaN | -3.888889 | NaN | NaN | 190601012000 | 190601 | 01 | 1906-01-01 20:00:00 | 1906-01-01 |
| 3 | 29440 | 190601020600 | NaN | 0.0 | NaN | 26.0 | NaN | NaN | -3.333333 | NaN | NaN | 190601020600 | 190601 | 01 | 1906-01-02 06:00:00 | 1906-01-01 |
| 4 | 29440 | 190601021300 | 270.0 | 7.0 | NaN | 27.0 | NaN | NaN | -2.777778 | NaN | NaN | 190601021300 | 190601 | 01 | 1906-01-02 13:00:00 | 1906-01-01 |
We have quite a few rows of weather data, and several observations per day. Our goal is to create an aggreated data frame that would have only one row per month.
To condense our data to monthly average values we can group our data based on the unique year and month combinations.
grouped = data.groupby("YEAR_MONTH")
Note
It is also possible to create combinations of years and months on-the-fly when grouping the data:
# Group the data
grouped = data.groupby(['YEAR', 'MONTH'])
Now, let’s explore the new variable grouped.
type(grouped)
pandas.core.groupby.generic.DataFrameGroupBy
len(grouped)
601
We have a new object with type DataFrameGroupBy with 601 groups. In order to understand what just happened, let’s also check the number of unique year and month combinations in our data.
data["YEAR_MONTH"].nunique()
601
Length of the grouped object should be the same as the number of unique values in the column we used for grouping. For each unique value, there is a group of data.
Let’s explore our grouped data even further.
We can check the “names” of each group.
# Next line will print out all 601 group "keys"
# This is commented out here to avoid excess output to the course website
# grouped.groups.keys()
Accessing data for one group#
Let us now check the contents for the group representing August 2019 (the name of that group is 201908. We can get the values of that hour from the grouped object using the get_group() method.
# Specify a month (as character string)
month = "201908"
# Select the group
group1 = grouped.get_group(month)
# Let's see what we have
group1
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | TEMP_C | MIN_C | MAX_C | TIME_STR | YEAR_MONTH | MONTH | DATE | YEAR_MONTH_DT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 753566 | 29440 | 201908010000 | 360.0 | 0.0 | NaN | 47.0 | NaN | NaN | 8.333333 | NaN | NaN | 201908010000 | 201908 | 08 | 2019-08-01 00:00:00 | 2019-08-01 |
| 753567 | 29440 | 201908010020 | NaN | 0.0 | NaN | 46.0 | NaN | NaN | 7.777778 | NaN | NaN | 201908010020 | 201908 | 08 | 2019-08-01 00:20:00 | 2019-08-01 |
| 753568 | 29440 | 201908010050 | NaN | 0.0 | NaN | 45.0 | NaN | NaN | 7.222222 | NaN | NaN | 201908010050 | 201908 | 08 | 2019-08-01 00:50:00 | 2019-08-01 |
| 753569 | 29440 | 201908010100 | 360.0 | 0.0 | NaN | 44.0 | NaN | NaN | 6.666667 | NaN | NaN | 201908010100 | 201908 | 08 | 2019-08-01 01:00:00 | 2019-08-01 |
| 753570 | 29440 | 201908010120 | NaN | 0.0 | NaN | 43.0 | NaN | NaN | 6.111111 | NaN | NaN | 201908010120 | 201908 | 08 | 2019-08-01 01:20:00 | 2019-08-01 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 755781 | 29440 | 201908312220 | 180.0 | 6.0 | NaN | 59.0 | NaN | NaN | 15.000000 | NaN | NaN | 201908312220 | 201908 | 08 | 2019-08-31 22:20:00 | 2019-08-01 |
| 755782 | 29440 | 201908312250 | 170.0 | 6.0 | NaN | 57.0 | NaN | NaN | 13.888889 | NaN | NaN | 201908312250 | 201908 | 08 | 2019-08-31 22:50:00 | 2019-08-01 |
| 755783 | 29440 | 201908312300 | 170.0 | 7.0 | NaN | 58.0 | NaN | NaN | 14.444444 | NaN | NaN | 201908312300 | 201908 | 08 | 2019-08-31 23:00:00 | 2019-08-01 |
| 755784 | 29440 | 201908312320 | 160.0 | 7.0 | NaN | 57.0 | NaN | NaN | 13.888889 | NaN | NaN | 201908312320 | 201908 | 08 | 2019-08-31 23:20:00 | 2019-08-01 |
| 755785 | 29440 | 201908312350 | 180.0 | 6.0 | NaN | 57.0 | NaN | NaN | 13.888889 | NaN | NaN | 201908312350 | 201908 | 08 | 2019-08-31 23:50:00 | 2019-08-01 |
2220 rows × 16 columns
Ahaa! As we can see, a single group contains a DataFrame with values only for that specific month and year. Let’s check the DataType of this group.
type(group1)
pandas.core.frame.DataFrame
So, as noted above, one group is a pandas DataFrame! This is really useful, because we can now use all the familiar DataFrame methods for calculating statistics, etc. for this specific group. We can, for example, calculate the average values for all variables using the statistical functions that we have seen already (e.g. mean, std, min, max, median, etc.).
We can do that by using the mean() function that we already did during Lesson 5.
Let’s calculate the mean for following attributes all at once:
DIRSPEEDGUSTTEMPTEMP_CMONTH
# Specify the columns that will be part of the calculation
mean_cols = ["DIR", "SPEED", "GUST", "TEMP_F", "TEMP_C"]
# Calculate the mean values all at one go
mean_values = group1[mean_cols].mean()
# Let's see what we have
print(mean_values)
DIR 320.335766
SPEED 6.769447
GUST 15.751678
TEMP_F 60.598649
TEMP_C 15.888138
dtype: float64
Above, we saw how you can access data from a single group. In order to get information about all groups (all months) we can use a for loop or methods available in the grouped object.
For loops and grouped objects#
When iterating over the groups in our DataFrameGroupBy object it is important to understand that a single group in our DataFrameGroupBy actually contains not only the actual values, but also information about the key that was used to do the grouping. Hence, when iterating over the data we need to assign the key and the values into separate variables.
So, let’s see how we can iterate over the groups and print the key and the data from a single group (again using break to only see what is happening for the first group).
# Iterate over groups
for key, group in grouped:
# Print key and group
print(f"Key:\n {key}")
print(f"\nFirst rows of data in this group:\n {group.head()}")
# Stop iteration with break command
break
Key:
190601
First rows of data in this group:
STATION_NUMBER TIME DIR SPEED GUST TEMP_F MAX MIN \
0 29440 190601010600 90.0 7.0 NaN 27.0 NaN NaN
1 29440 190601011300 NaN 0.0 NaN 27.0 NaN NaN
2 29440 190601012000 NaN 0.0 NaN 25.0 NaN NaN
3 29440 190601020600 NaN 0.0 NaN 26.0 NaN NaN
4 29440 190601021300 270.0 7.0 NaN 27.0 NaN NaN
TEMP_C MIN_C MAX_C TIME_STR YEAR_MONTH MONTH DATE \
0 -2.777778 NaN NaN 190601010600 190601 01 1906-01-01 06:00:00
1 -2.777778 NaN NaN 190601011300 190601 01 1906-01-01 13:00:00
2 -3.888889 NaN NaN 190601012000 190601 01 1906-01-01 20:00:00
3 -3.333333 NaN NaN 190601020600 190601 01 1906-01-02 06:00:00
4 -2.777778 NaN NaN 190601021300 190601 01 1906-01-02 13:00:00
YEAR_MONTH_DT
0 1906-01-01
1 1906-01-01
2 1906-01-01
3 1906-01-01
4 1906-01-01
OK, so from here we can see that the key contains the name of the group YEARMO.
Let’s build on this and see how we can create a DataFrame where we calculate the mean values for all those weather attributes that we were interested in. We will repeat some of the earlier steps here so you can see and better understand what is happening.
# Create an empty DataFrame for the aggregated values
monthly_data = pd.DataFrame()
# The columns that we want to aggregate
mean_cols = ["DIR", "SPEED", "GUST", "TEMP_F", "TEMP_C"]
# Iterate over the groups
for key, group in grouped:
# Calculate mean
mean_values = group[mean_cols].mean()
# Add the ´key´ (i.e. the date+time information) into the aggregated values
mean_values["YEAR_MONTH"] = key
# Convert the mean_values series to a DataFrame and make it have a row orientation
row = mean_values.to_frame().transpose()
# Concatenate the aggregated values into the monthly_data DataFrame
monthly_data = pd.concat([monthly_data, row], ignore_index=True)
Note
You can find a slightly more efficient approach for this same kind of aggregation in Chapter 3 of the Python for Geographic Data Analysis textbook.
Now, let us see what we have.
print(monthly_data)
DIR SPEED GUST TEMP_F TEMP_C YEAR_MONTH
0 218.181818 13.204301 NaN 25.526882 -3.596177 190601
1 178.095238 13.142857 NaN 25.797619 -3.445767 190602
2 232.043011 15.021505 NaN 22.806452 -5.107527 190603
3 232.045455 13.811111 NaN 38.822222 3.790123 190604
4 192.820513 10.333333 NaN 55.526882 13.07049 190605
.. ... ... ... ... ... ...
596 370.992008 8.13849 17.251852 61.7434 16.524111 201906
597 294.433641 5.785714 15.034722 61.569955 16.427753 201907
598 320.335766 6.769447 15.751678 60.598649 15.888138 201908
599 306.491058 6.363594 15.173285 49.958137 9.976743 201909
600 239.577465 10.169014 17.470588 42.774648 5.985915 201910
[601 rows x 6 columns]
Awesome! Now we have aggregated our data and we have a new DataFrame called monthly_data where we have mean values for each month in the data set.
Finding the mean for all groups at once#
We can also achieve the same result by computing the mean of all columns for all groups in the grouped object.
grouped.mean()
| STATION_NUMBER | TIME | DIR | SPEED | GUST | TEMP_F | MAX | MIN | TEMP_C | MIN_C | MAX_C | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| YEAR_MONTH | |||||||||||
| 190601 | 29440.0 | 1.906012e+11 | 218.181818 | 13.204301 | NaN | 25.526882 | NaN | NaN | -3.596177 | NaN | NaN |
| 190602 | 29440.0 | 1.906021e+11 | 178.095238 | 13.142857 | NaN | 25.797619 | NaN | NaN | -3.445767 | NaN | NaN |
| 190603 | 29440.0 | 1.906032e+11 | 232.043011 | 15.021505 | NaN | 22.806452 | NaN | NaN | -5.107527 | NaN | NaN |
| 190604 | 29440.0 | 1.906042e+11 | 232.045455 | 13.811111 | NaN | 38.822222 | NaN | NaN | 3.790123 | NaN | NaN |
| 190605 | 29440.0 | 1.906052e+11 | 192.820513 | 10.333333 | NaN | 55.526882 | NaN | NaN | 13.070490 | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 201906 | 29440.0 | 2.019062e+11 | 370.992008 | 8.138490 | 17.251852 | 61.743400 | 67.316667 | 55.600000 | 16.524111 | 13.111111 | 19.620370 |
| 201907 | 29440.0 | 2.019072e+11 | 294.433641 | 5.785714 | 15.034722 | 61.569955 | 67.774194 | 55.903226 | 16.427753 | 13.279570 | 19.874552 |
| 201908 | 29440.0 | 2.019082e+11 | 320.335766 | 6.769447 | 15.751678 | 60.598649 | 65.935484 | 55.016129 | 15.888138 | 12.786738 | 18.853047 |
| 201909 | 29440.0 | 2.019092e+11 | 306.491058 | 6.363594 | 15.173285 | 49.958137 | 53.766667 | 45.350000 | 9.976743 | 7.416667 | 12.092593 |
| 201910 | 29440.0 | 2.019100e+11 | 239.577465 | 10.169014 | 17.470588 | 42.774648 | 48.500000 | 41.000000 | 5.985915 | 5.000000 | 9.166667 |
601 rows × 11 columns
Detecting warm months#
Now that we have aggregated our data on monthly level, all we need to do is to sort our results in order to check which years had the warmest April temperatures. A simple approach is to select all Aprils from the data, group the data and check which group(s) have the highest mean value.
We can start this by selecting all records that are from April (regardless of the year).
aprils = data[data["MONTH"] == "04"]
Next, we can take a subset of columns that might contain interesting information.
aprils = aprils[["STATION_NUMBER", "TEMP_F", "TEMP_C", "YEAR_MONTH"]]
We can group by year and month.
grouped = aprils.groupby(by="YEAR_MONTH")
And then we can calculate the mean for each group.
monthly_mean = grouped.mean()
monthly_mean.head()
| STATION_NUMBER | TEMP_F | TEMP_C | |
|---|---|---|---|
| YEAR_MONTH | |||
| 190604 | 29440.0 | 38.822222 | 3.790123 |
| 190704 | 29440.0 | 36.111111 | 2.283951 |
| 190804 | 29440.0 | 36.811111 | 2.672840 |
| 190904 | 29440.0 | 31.977778 | -0.012346 |
| 191004 | 29440.0 | 39.833333 | 4.351852 |
Finally, we can sort and check the highest temperature values. We can sort the data frame in a descending order to do this.
monthly_mean.sort_values(by="TEMP_C", ascending=False).head(10)
| STATION_NUMBER | TEMP_F | TEMP_C | |
|---|---|---|---|
| YEAR_MONTH | |||
| 201904 | 29440.0 | 42.472030 | 5.817794 |
| 199004 | 29440.0 | 41.918084 | 5.510047 |
| 198904 | 29440.0 | 41.369647 | 5.205360 |
| 201104 | 29440.0 | 41.290730 | 5.161517 |
| 200404 | 29440.0 | 41.249676 | 5.138709 |
| 200204 | 29440.0 | 41.132353 | 5.073529 |
| 198304 | 29440.0 | 41.016183 | 5.008991 |
| 200804 | 29440.0 | 40.962343 | 4.979079 |
| 200004 | 29440.0 | 40.777778 | 4.876543 |
| 199904 | 29440.0 | 40.695291 | 4.830717 |
So, how did April 2019 rank at the Tampere Pirkkala observation station?
Repeating the data analysis with a larger dataset#
To wrap up today’s lesson, let’s repeat the data analysis steps above for all the available data we have (!). First, it would be good to confirm the path to the directory where all the input data are located.
The idea is, that we will repeat the analysis process for each input file using a (rather long) for loop! Here we have all the main analysis steps with some additional output info, all in one long code cell.
# Read selected columns of data using varying amount of spaces as separator and specifying * characters as NoData values
data = pd.read_csv(
fp,
delim_whitespace=True,
usecols=["USAF", "YR--MODAHRMN", "DIR", "SPD", "GUS", "TEMP", "MAX", "MIN"],
na_values=["*", "**", "***", "****", "*****", "******"],
)
# Rename the columns
new_names = {
"USAF": "STATION_NUMBER",
"YR--MODAHRMN": "TIME",
"SPD": "SPEED",
"GUS": "GUST",
"TEMP": "TEMP_F",
}
data = data.rename(columns=new_names)
# Print info about the current input file:
print(f"STATION NUMBER: {data.at[0, 'STATION_NUMBER']}")
print(f"NUMBER OF OBSERVATIONS: {len(data)}")
# Create column
col_name = "TEMP_C"
data[col_name] = None
# Convert tempetarues from Fahrenheits to Celsius
data["TEMP_C"] = data["TEMP_F"].apply(fahr_to_celsius)
# Convert TIME to string
data["TIME_STR"] = data["TIME"].astype(str)
# Parse year and month
data["MONTH"] = data["TIME_STR"].str.slice(start=5, stop=6).astype(int)
data["YEAR"] = data["TIME_STR"].str.slice(start=0, stop=4).astype(int)
# Extract observations for the months of April
aprils = data[data["MONTH"] == 4]
# Take a subset of columns
aprils = aprils[["STATION_NUMBER", "TEMP_F", "TEMP_C", "YEAR", "MONTH"]]
# Group by year and month
grouped = aprils.groupby(by=["YEAR", "MONTH"])
# Get mean values for each group
monthly_mean = grouped.mean()
# Print info
print(monthly_mean.sort_values(by="TEMP_C", ascending=False).head(5))
print("\n")
STATION NUMBER: 29440
NUMBER OF OBSERVATIONS: 757983
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
2019 4 29440.0 42.472030 5.817794
1990 4 29440.0 41.918084 5.510047
1989 4 29440.0 41.369647 5.205360
2011 4 29440.0 41.290730 5.161517
2004 4 29440.0 41.249676 5.138709
print(data.at[0, "STATION_NUMBER"])
29440
At this point we will use the glob() function from the module glob to list our input files. glob is a handy function for finding files in a directrory that match a given pattern, for example.
import glob
file_list = glob.glob(r"/home/jovyan/shared/data/L6/0*txt")
Show code cell content
# IGNORE THIS CELL
# It is only needed to generate the course website
file_list = glob.glob(r"data/0*txt")
Note
Note that we’re using the * character as a wildcard, so any file that starts with data/0 and ends with txt will be added to the list of files we will iterate over. We specifically use data/0 as the starting part of the file names to avoid having our metadata files included in the list!
Note
If you are using Jupyter Lab installed on your own computer, then the file_list variable you should use is defined below:
file_list = glob.glob(r"data/0*txt")
print(f"Number of files in the list: {len(file_list)}")
print(file_list)
Number of files in the list: 15
['data/029170.txt', 'data/028690.txt', 'data/029820.txt', 'data/029700.txt', 'data/028970.txt', 'data/029070.txt', 'data/029500.txt', 'data/029110.txt', 'data/028750.txt', 'data/029720.txt', 'data/029440.txt', 'data/028360.txt', 'data/029810.txt', 'data/029740.txt', 'data/029350.txt']
Now, you should have all the relevant file names in a list, and we can loop over the list using a for loop.
for fp in file_list:
print(fp)
data/029170.txt
data/028690.txt
data/029820.txt
data/029700.txt
data/028970.txt
data/029070.txt
data/029500.txt
data/029110.txt
data/028750.txt
data/029720.txt
data/029440.txt
data/028360.txt
data/029810.txt
data/029740.txt
data/029350.txt
# Repeat the analysis steps for each input file:
for fp in file_list:
# Read selected columns of data using varying amount of spaces as separator and specifying * characters as NoData values
data = pd.read_csv(
fp,
delim_whitespace=True,
usecols=["USAF", "YR--MODAHRMN", "DIR", "SPD", "GUS", "TEMP", "MAX", "MIN"],
na_values=["*", "**", "***", "****", "*****", "******"],
)
# Rename the columns
new_names = {
"USAF": "STATION_NUMBER",
"YR--MODAHRMN": "TIME",
"SPD": "SPEED",
"GUS": "GUST",
"TEMP": "TEMP_F",
}
data = data.rename(columns=new_names)
# Print info about the current input file:
print(f"STATION NUMBER: {data.at[0, 'STATION_NUMBER']}")
print(f"NUMBER OF OBSERVATIONS: {len(data)}")
# Create column
col_name = "TEMP_C"
data[col_name] = None
# Convert tempetarues from Fahrenheits to Celsius
data["TEMP_C"] = data["TEMP_F"].apply(fahr_to_celsius)
# Convert TIME to string
data["TIME_STR"] = data["TIME"].astype(str)
# Parse year and month
data["MONTH"] = data["TIME_STR"].str.slice(start=5, stop=6).astype(int)
data["YEAR"] = data["TIME_STR"].str.slice(start=0, stop=4).astype(int)
# Extract observations for the months of April
aprils = data[data["MONTH"] == 4]
# Take a subset of columns
aprils = aprils[["STATION_NUMBER", "TEMP_F", "TEMP_C", "YEAR", "MONTH"]]
# Group by year and month
grouped = aprils.groupby(by=["YEAR", "MONTH"])
# Get mean values for each group
monthly_mean = grouped.mean()
# Print info
print(monthly_mean.sort_values(by="TEMP_C", ascending=False).head(5))
print("\n")
STATION NUMBER: 29170
NUMBER OF OBSERVATIONS: 561097
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
1937 4 29170.0 43.289157 6.271754
2019 4 29170.0 40.666820 4.814900
2011 4 29170.0 40.015962 4.453312
2001 4 29170.0 39.713228 4.285126
1906 4 29170.0 39.688889 4.271605
STATION NUMBER: 28690
NUMBER OF OBSERVATIONS: 542788
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
2011 4 28690.0 35.430640 1.905911
2019 4 28690.0 35.215114 1.786174
2016 4 28690.0 35.031103 1.683946
1989 4 28690.0 34.612766 1.451537
2002 4 28690.0 34.279855 1.266586
STATION NUMBER: 29820
NUMBER OF OBSERVATIONS: 198334
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
2019 4 29820.0 41.182197 5.101221
1990 4 29820.0 41.144681 5.080378
2014 4 29820.0 40.497908 4.721060
2008 4 29820.0 39.941423 4.411901
1913 4 29820.0 39.622222 4.234568
STATION NUMBER: 29700
NUMBER OF OBSERVATIONS: 473881
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
1921 4 29700.0 42.811111 6.006173
2000 4 29700.0 42.375587 5.764215
1990 4 29700.0 42.054167 5.585648
2019 4 29700.0 41.548747 5.304859
2004 4 29700.0 41.493392 5.274107
STATION NUMBER: 28970
NUMBER OF OBSERVATIONS: 555740
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
1921 4 28970.0 41.688889 5.382716
1999 4 28970.0 39.073600 3.929778
2019 4 28970.0 38.706456 3.725809
1989 4 28970.0 38.362869 3.534927
2011 4 28970.0 38.094172 3.385651
STATION NUMBER: 29070
NUMBER OF OBSERVATIONS: 83567
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
2014 4 29070.0 35.437326 1.909626
2015 4 29070.0 34.437209 1.354005
2004 4 29070.0 34.347032 1.303907
2016 4 29070.0 34.303199 1.279555
2008 4 29070.0 34.241667 1.245370
STATION NUMBER: 29500
NUMBER OF OBSERVATIONS: 103105
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
2019 4 29500.0 41.639777 5.355432
2008 4 29500.0 40.838936 4.910520
2014 4 29500.0 40.226415 4.570231
2016 4 29500.0 39.176634 3.987019
2011 4 29500.0 38.647826 3.693237
STATION NUMBER: 29110
NUMBER OF OBSERVATIONS: 483784
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
1921 4 29110.0 42.166667 5.648148
2004 4 29110.0 41.682699 5.379277
1989 4 29110.0 41.420168 5.233427
1937 4 29110.0 40.671429 4.817460
2019 4 29110.0 40.636300 4.797945
STATION NUMBER: 28750
NUMBER OF OBSERVATIONS: 474562
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
1989 4 28750.0 39.008403 3.893557
1983 4 28750.0 38.758475 3.754708
2019 4 28750.0 38.651599 3.695333
2002 4 28750.0 38.270419 3.483566
1994 4 28750.0 38.145833 3.414352
STATION NUMBER: 29720
NUMBER OF OBSERVATIONS: 843688
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
2019 4 29720.0 43.558414 6.421341
1990 4 29720.0 43.313576 6.285320
2000 4 29720.0 42.663169 5.923983
2008 4 29720.0 42.349642 5.749801
2004 4 29720.0 41.903492 5.501940
STATION NUMBER: 29440
NUMBER OF OBSERVATIONS: 757983
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
2019 4 29440.0 42.472030 5.817794
1990 4 29440.0 41.918084 5.510047
1989 4 29440.0 41.369647 5.205360
2011 4 29440.0 41.290730 5.161517
2004 4 29440.0 41.249676 5.138709
STATION NUMBER: 28360
NUMBER OF OBSERVATIONS: 193825
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
1937 4 28360.0 38.738095 3.743386
2011 4 28360.0 36.699571 2.610873
1921 4 28360.0 36.622222 2.567901
2002 4 28360.0 36.500000 2.500000
2019 4 28360.0 34.979138 1.655076
STATION NUMBER: 29810
NUMBER OF OBSERVATIONS: 199330
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
1990 4 29810.0 41.157895 5.087719
2019 4 29810.0 40.783032 4.879462
2014 4 29810.0 40.058036 4.476687
2008 4 29810.0 40.044881 4.469378
2016 4 29810.0 39.270308 4.039060
STATION NUMBER: 29740
NUMBER OF OBSERVATIONS: 931767
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
2000 4 29740.0 43.479793 6.377663
2019 4 29740.0 43.464070 6.368928
1990 4 29740.0 43.375078 6.319488
2008 4 29740.0 43.341429 6.300794
2011 4 29740.0 42.750702 5.972612
STATION NUMBER: 29350
NUMBER OF OBSERVATIONS: 559667
STATION_NUMBER TEMP_F TEMP_C
YEAR MONTH
1921 4 29350.0 45.144444 7.302469
1925 4 29350.0 40.777778 4.876543
2011 4 29350.0 40.670108 4.816727
2019 4 29350.0 40.585002 4.769446
2001 4 29350.0 39.662827 4.257126
So, what can we conclude about how warm April 2019 was in Finland? Was it actually the warmest April on record? If so, in which stations?


